Order Guide
Related Products
Related Services
Related Reviews
rtStar™ Pre-designed tRF&tiRNA Primer Sets (H/M)
Benefits
• Reliable results with full validated primers
• High sensitivity and specificity
• Accurate quantification over a wide linear range
• Time and cost savings with validated primers and SYBR Green assay
The Primer Sets perform superbly with the rtStar™ First-Strand cDNA Synthesis Kit (3’ and 5’ adaptor) (CAT# AS-FS-003) and Arraystar SYBR Green qPCR Master Mix.
| Product Name | Catalog No. | Size | Price |
|---|---|---|---|
| rtStar™ Pre-designed Human tRF&tiRNA qPCR Primer Pair | AS-NR-002-1-M | 200 reactions | $175.00 |
| rtStar™ Pre-designed Mouse tRF qPCR Primer Pair | AS-NR-002M-1-M | 200 reactions | $175.00 |
rtStar™ Pre-designed Human tRF&tiRNA Primer Sets are part of the nrStar™ tRF&tiRNA PCR System, and can be used in combination with rtStar™ First-Strand cDNA Synthesis Kit (3’ and 5’ adaptor)(AS-FS-003) for tRF&tiRNA detection and quantification. All the primers have been validated in numerous sample types, and guarantee the most accurate and specific performance when used with Arraystar SYBR Green qPCR Master Mix (Figure 1).
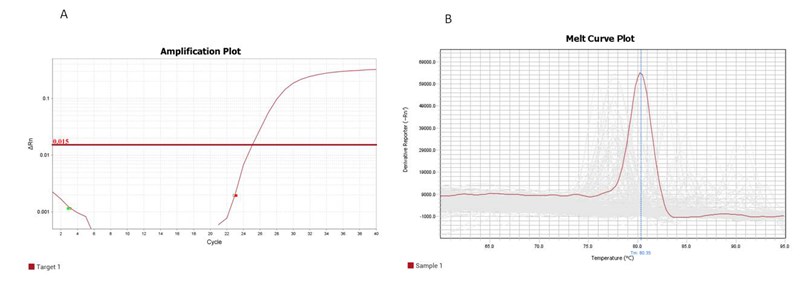
Figure 1. Amplification curve (A) and Dissociation curve (B) of example Human tRF-5005A.
| tRF&tiRNA name | tRF sequence | Source tRNA | Catalog Number |
| 3’tiR_007_GluTTC (n) | TTTCACCCAGGCGGCCCGGGTTCGACTCCCGGTGTGGGAACCA | GluTTC (n) | AS-NR-002-1-001 |
| 3’tiR_012_ArgCCT (n) | CCTAAGCCAGGGATTGTGGGTTCGAGTCCCACCCGGGGTACCA | ArgCCT (n) | AS-NR-002-1-002 |
| 3’tiR_026_GlnCTG (n) | TGAATCCAGCGATCCGAGTTCAAATCTCGGTGGAACCTCCA | GlnCTG (n) | AS-NR-002-1-003 |
| 3’tiR_028_HisGTG (mt) | TGTGAATCTGACAACAGAGGCTTACGACCCCTTATTTACCC | HisGTG (mt) | AS-NR-002-1-004 |
| 3’tiR_037_ArgCCG (n) | GGAGCTGGGGATTGTGGGTTCGAGTCCCATCTGGGTCGCCA | ArgCCG (n) | AS-NR-002-1-005 |
| 3’tiR_056_ValTAC (mt) | TACACTTAGGAGATTTCAACTTAACTTGACCGCTCTGACCA | ValTAC (mt) | AS-NR-002-1-006 |
| 3’tiR_060_MetCAT (n) | TCATAATCTGAAGGTCGTGAGTTCGATCCTCACACGGGGCACCA | MetCAT (n) | AS-NR-002-1-007 |
| 3’tiR_063_ArgCCT (n) | TCCTAAGCCAGGGATTGTGGGTTCGAGTCCCATCTGGGGTGCCA | ArgCCT (n) | AS-NR-002-1-008 |
| 3’tiR_075_GluTTC (mt) GluTTC (mt-la) |
TTCATATCATTGGTCGTGGTTGTAGTCCGTGCGAGAATACC | GluTTC (mt) GluTTC (mt-la) |
AS-NR-002-1-009 |
| 3’tiR_078_ArgTCT (n) | TTCTAATTCAAAGGTTCCGGGTTCGAGTCCCGGCGGAGTCGCCA | ArgTCT (n) | AS-NR-002-1-010 |
| 3’tiR_080_ProTGG (mt) | TTGGGTGCTAATGGTGGAGTTAAAGACTTTTTCTCTGACC | ProTGG (mt) | AS-NR-002-1-011 |
| 3’tiR_082_ThrTGT (mt) | TTGTAAACCGGAGATGAAAACCTTTTTCCAAGGACACCA | ThrTGT (mt) | AS-NR-002-1-012 |
| 3’tiR_088_LysCTT (n) | CTTAATCTCAGGGTCGTGGGTTCGAGCCCCACGTTGGGCGCCA | LysCTT (n) | AS-NR-002-1-013 |
| 5003/4C | GCATGGGTGGTTCAGTGGTAGAATTCTCGCC GCATTGGTGGTTCAGTGGTAGAATTCTCGCC |
GlyGCC GlyGCC |
AS-NR-002-1-014 |
| 5008C | GCGTTGGTGGTATAGTGGTGAGCATAGCTG | GlyTCC | AS-NR-002-1-015 |
| 5009C | GCTTCTGTAGTGTAGTGGTTATCACGTTCGC | ValCAC | AS-NR-002-1-016 |
| 5016C | GGGGGTATAGCTCAGTGGTAGAGCATTTGACT | CysGCA | AS-NR-002-1-017 |
| 5026/27C | GTTTCCGTAGTGTAGTGGTCATCACGTTCGC GTTTCCGTAGTGTAGTGGTTATCACGTTCGC |
ValAAC ValAAC ValCAC |
AS-NR-002-1-018 |
| TRF62 | AGCAGAGTGGCGCAGCGGAAGCGTGCTGG | MetCAT | AS-NR-002-1-019 |
| TRF315 | GCCCGGCTAGCTCAGTCGGTAGAGCATGG | LysCTT | AS-NR-002-1-020 |
| TRF327 | GCCTTGGTGGTGCAGTGGTAGAATTCTCGCCT | GlyCCC | AS-NR-002-1-021 |
| TRF353 | GGCCCCATGGTGTAATGGTTAGCACTCTGGA | GlnTTG | AS-NR-002-1-022 |
| TRF419 | GGTAGTGTGGCCGAGCGGTCTAAGGCGCTG | LeuTAG | AS-NR-002-1-023 |
| tiRNA-5033-ProTGG-1 | GGCTCGTTGGTCTAGGGGTATGATTCTCGGTTT | ProTGG | AS-NR-002-1-024 |
| tiRNA-5033-GluTTC-1 | TCCCACATGGTCTAGCGGTTAGGATTCCTGGTT | GluTTC | AS-NR-002-1-025 |
| tiRNA-5029-GlyGCC-2 | GCGCCGCTGGTGTAGTGGTATCATGCAAG | GlyGCC | AS-NR-002-1-026 |
| tiRNA-5034-ValCAC-2 | GCTTCTGTAGTGTAGTGGTTATCACGTTCGCCTC | ValCAC | AS-NR-002-1-027 |
| tiRNA-5034-ValCAC-3 | GTTTCCGTAGTGTAGCGGTTATCACATTCGCCTC | ValCAC | AS-NR-002-1-028 |
| tiRNA-5030-HisGTG-1 | GCCGTGATCGTATAGTGGTTAGTACTCTGC | HisGTG | AS-NR-002-1-029 |
| tiRNA-5030-GluTTC-1 | TCCCACATGGTCTAGCGGTTAGGATTCCTG | GluTTC | AS-NR-002-1-030 |
| tiRNA-5033-GluTTC-2 | TCCCATATGGTCTAGCGGTTAGGATTCCTGGTT | GluTTC | AS-NR-002-1-031 |
| tiRNA-5030-LysCTT-2 | GCCCGGCTAGCTCAGTCGGTAGAGCATGGG | LysCTT | AS-NR-002-1-032 |
| tiRNA-5029-ProAGG | GGCTCGTTGGTCTAGGGGTATGATTCTCG | ProAGG | AS-NR-002-1-033 |
| tiRNA-5031-GluTTC-1 | TCCCTGGTGGTCTAGTGGCTAGGATTCGGCG | GluTTC | AS-NR-002-1-034 |
| tiRNA-5031-HisGTG-1 | GCCGTGATCGTATAGTGGTTAGTACTCTGCG | HisGTG | AS-NR-002-1-035 |
| tiRNA-5031-GluCTC-1 | TCCCTGGTGGTCTAGTGGTTAGGATTCGGCG | GluCTC | AS-NR-002-1-036 |
| tiRNA-5034-GlyCCC-1 | GCGCCGCTGGTGTAGTGGTATCATGCAAGATTCC | GlyCCC | AS-NR-002-1-037 |
| tiRNA-5034-GluTTC-1 | TCCCACATGGTCTAGCGGTTAGGATTCCTGGTTT | GluTTC | AS-NR-002-1-038 |
| tiRNA-5033-LysTTT-1 | GCCCGGATAGCTCAGTCGGTAGAGCATCAGACT | LysTTT | AS-NR-002-1-039 |
| tiRNA-5032-LysTTT-1 | GCCCGGATAGCTCAGTCGGTAGAGCATCAGAC | LysTTT | AS-NR-002-1-040 |
| tiRNA-5031-PheGAA | GCCGAAATAGCTCAGTTGGGAGAGCTTTAGA | PheGAA | AS-NR-002-1-041 |
| tiRNA-5034-ValTAC-3 | GGTTCCATAGTGTAGTGGTTATCACATCTGCTTT | ValTAC | AS-NR-002-1-042 |
| tiRNA-5030-GlnTTG-3 | GGTCCCGTGGTGTAATGGTTAGCACTCTGG | GlnTTG | AS-NR-002-1-043 |
| tiRNA-5029-GlyGCC-3 | GCATTGGTGGTTCAGTGGTAGAATTCTCG | GlyGCC | AS-NR-002-1-044 |
| tiRNA-5035-GluTTC-1 | TCCCACATGGTCTAGCGGTTAGGATTCCTGGTTTT | GluTTC | AS-NR-002-1-045 |
| tiRNA-5035-GluTTC-2 | TCCCATATGGTCTAGCGGTTAGGATTCCTGGTTTT | GluTTC | AS-NR-002-1-046 |
| tiRNA-5034-GluTTC-2 | TCCCATATGGTCTAGCGGTTAGGATTCCTGGTTT | GluTTC | AS-NR-002-1-047 |
| tiRNA-5035-GluTTC-3 | TCCCTGGTGGTCTAGTGGCTAGGATTCGGCGCTTT | GluTTC | AS-NR-002-1-048 |
| tiRNA-5032-GluTTC-1 | TCCCACATGGTCTAGCGGTTAGGATTCCTGGT | GluTTC | AS-NR-002-1-049 |
| tiRNA-5035-GluCTC | TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCTCT | GluCTC | AS-NR-002-1-050 |
| tiRNA-5030-SerGCT-3 | GACGAGGTGGCCGAGTGGTTAAGGCGATGG | SerGCT | AS-NR-002-1-051 |
| tiRNA-5030-SerGCT-1 | GACGAGGTGGCCGAGTGGTTAAGGCAATGG | SerGCT | AS-NR-002-1-052 |
| tiRNA-5031-HisGTG-2 | GCCGTAATCGTATAGTGGTTAGTACTCTGCG | HisGTG | AS-NR-002-1-053 |
| tiRNA-5029-AlaAGC-1 | GGGGGATTAGCTCAAATGGTAGAGCCCTC | AlaAGC | AS-NR-002-1-054 |
| tiRNA-5032-LysCTT-1 | GCCCGGCTAGCTCAGTCGGTAGAGCATGGGAC | LysCTT | AS-NR-002-1-055 |
| 1001 | GAAGCGGGTGCTCTTATTTT | SerTGA | AS-NR-002-1-056 |
| 1003 | GCTAAGGAAGTCCTGTGCTCAGTTTT | SerGCT | AS-NR-002-1-057 |
| 1004 | GTGTGTAGCTGCACTTTT | AspGTC | AS-NR-002-1-058 |
| 1005 | ATGTGGTGGCTTACTTT | SerGCT | AS-NR-002-1-059 |
| 1006 | GTGTAAGCAGGGTCGTTTT | ArgACG | AS-NR-002-1-060 |
| 1007 | TTCAAAGGTGAACGTTT | GlnTTG | AS-NR-002-1-061 |
| 1010 | GGTGTGGTCTGTTGTTT | ValTAC | AS-NR-002-1-062 |
| 1012 | GCACGAAAATGTGTTTT | GlyGCC | AS-NR-002-1-063 |
| 1013 | GGCGATCACGTAGATTTT | AlaCGC | AS-NR-002-1-064 |
| 1015 | TGTGCTCCGGAGTTACCTCGTTT | CysGCA | AS-NR-002-1-065 |
| 1020 | GAGAGCGCTCGGTTTTT | PheGAA | AS-NR-002-1-066 |
| 1025 | GAGGGTTCTCACCTTCTCTCTCCGATTT | IleAAT | AS-NR-002-1-067 |
| 1026 | TTCCGTGGGTTTGTTTT | IleAAT | AS-NR-002-1-068 |
| 1027 | ATGGCCGCATATATTT | MetCAT | AS-NR-002-1-069 |
| 1028 | GAGGCTTAAACTTTT | AspGTC | AS-NR-002-1-070 |
| 1029 | ATAGGTATTAAGGTTTT | AlaTGC | AS-NR-002-1-071 |
| 1030 | GGAATGTCAGCTTTT | SerAGA | AS-NR-002-1-072 |
| 1031 | ACAAGTGCGGTTTTTT | TyrGTA | AS-NR-002-1-073 |
| 1032 | AAGAGGAGTTGTTTT | LeuTAA | AS-NR-002-1-074 |
| 1033 | GTGGGGTGCCTCACAGCTTCGCTGCGTGAGCATTTT | ArgACG | AS-NR-002-1-075 |
| 1035 | GATATCCAACCTTCGGCTATAGGGTGGAGACTTTTT | ThrCGT | AS-NR-002-1-076 |
| 1036 | GGGTTGCTGTCTTTT | LeuAAG | AS-NR-002-1-077 |
| 1037 | ACCTCAGAAGGTCTCACTTT | LeuTAG | AS-NR-002-1-078 |
| 1038 | AGGTGAAAGTTCCTTT | ArgCCT | AS-NR-002-1-079 |
| 1039 | TCGAGAGGGGCTGTGCTCGCAAGGTTTCTTT | ArgCCT | AS-NR-002-1-080 |
| 1040 | GTGGGTGGCTTTTTT | IleAAT | AS-NR-002-1-081 |
| 1041 | TGCGGTACCACTTTT | GlyTCC | AS-NR-002-1-082 |
| 1042 | AACCGAGCGTCCAAGCTCTTTCCATTTT | ThrAGT | AS-NR-002-1-083 |
| 3002B | TCAAATCCCGGACGAGCCCCCA | ProCGG | AS-NR-002-1-084 |
| 3004B | TCAAATCTCGGTGGGACCTCCA | GlnTTG | AS-NR-002-1-085 |
| 3006B | TCAAGTCCCTGTTCGGGCGCCA | LysTTT | AS-NR-002-1-086 |
| 3031B | TCGCTGGTTCGAATCCGGCTCGGAGGACCA | TyrGTA | AS-NR-002-1-087 |
| 3033A | CCCACCCAGGGACGCCA | AsnGTT | AS-NR-002-1-088 |
| 3030A | TTCCGGCTCGAAGGACCA | TyrGTA | AS-NR-002-1-089 |
| 3030B | TCGATTCCGGCTCGAAGGACCA | TyrGTA | AS-NR-002-1-090 |
| 3002A | ATCCCGGACGAGCCCCCA | ProAGG | AS-NR-002-1-091 |
| 3003A | TCCGGGTGCCCCCTCCA | CysGCA | AS-NR-002-1-092 |
| 3003B | TCAAATCCGGGTGCCCCCTCCA | CysGCA | AS-NR-002-1-093 |
| 3006A | TCCCTGTTCGGGCGCCA | LysTTT | AS-NR-002-1-094 |
| 3008A | ACCGGGCGGAAACACCA | ValAAC | AS-NR-002-1-095 |
| 3008B | TCGAAACCGGGCGGAAACACCA | ValAAC | AS-NR-002-1-096 |
| 3009B | TCGAACCCCACTCCTGGTACCA | LeuTAA | AS-NR-002-1-097 |
| 3011/12A | ATCCCACTCCTGACACCA ATCCCACTTCTGACACCA |
LeuCAG LeuCAA LeuCAG |
AS-NR-002-1-098 |
| 3016/18/22B | TCGAGCCCCACGTTGGGCGCCA TCGAGCCTCAGAGAGGGCACCA |
LysCTT MetCAT |
AS-NR-002-1-099 |
| 3017A | AGCCCCAGTGGAACCACCA | ValTAC | AS-NR-002-1-100 |
| 3017B | TCGAGCCCCAGTGGAACCACCA | ValTAC | AS-NR-002-1-101 |
| 3019/20/21B | TCGATCCCCAGTACCTCCACCA TCGATCCCCGGCACCTCCACCA TCGATCCCCGGCATCTCCACCA |
AlaAGC AlaTGC AlaCGC AlaTGC |
AS-NR-002-1-102 |
| 3026B | TCGATTCCCGGCCAACGCACCA | GlyTCC | AS-NR-002-1-103 |
| 3027/28B | TCGATTCCCGGCCCATGCACCA TCGATTCCCGGCCAATGCACCA |
GlyGCC GlyGCC |
AS-NR-002-1-104 |
| 3019A | TCCCCAGTACCTCCACCA | AlaAGC | AS-NR-002-1-105 |
| 3020/21A | TCCCCGGCACCTCCACCA TCCCCGGCATCTCCACCA |
AlaTGC AlaAGC AlaTGC AlaCGC |
AS-NR-002-1-106 |
| 3022A | TCCCCGTACGGGCCACCA | IleAAT | AS-NR-002-1-107 |
| 3026/27/28A | TTCCCGGCCAACGCACCA TCCCGGCCAATGCACCA TCCCGGCCCATGCACCA |
GlyTCC GlyCCC GlyGCC GlyGCC |
AS-NR-002-1-108 |
| 3029A | TTCCCGGTCAGGGAACCA | GluCTC | AS-NR-002-1-109 |
| 3031A | TCCGGCTCGGAGGACCA | TyrGTA | AS-NR-002-1-110 |
| 5001A | CCTTCGATAGCTCAG | TyrGTA | AS-NR-002-1-111 |
| 5001B | CCTTCGATAGCTCAGCTGGTAGAGC | TyrGTA | AS-NR-002-1-112 |
| 5002A | GACCCAGTGGCCTA | ArgCCG | AS-NR-002-1-113 |
| 5002B | GACCCAGTGGCCTAATGGA | ArgCCG | AS-NR-002-1-114 |
| 5008B | GCGTTGGTGGTATAGTGGTGAGC | GlyTCC | AS-NR-002-1-115 |
| 5009A | GCTTCTGTAGTGTAG | ValCAC | AS-NR-002-1-116 |
| 5009B | GCTTCTGTAGTGTAGTGGTTATC | ValCAC | AS-NR-002-1-117 |
| 5010A | GGCCGGTTAGCTCAG | SerACT | AS-NR-002-1-118 |
| 5011A | GGCTCGTTGGTCTAG | ProCGG | AS-NR-002-1-119 |
| 5012B | GGCTCGTTGGTCTAGGGGTATGA | ProCGG | AS-NR-002-1-120 |
| 5013B | GGCTCGTTGGTCTAGGGGTAT | ProCGG | AS-NR-002-1-121 |
| 5015/17A | GGGGGTATAGCTC GGGGGTGTAGCTC |
AlaAGC TyrGTA ValAAC CysGCA AlaTGC AlaCGC AlaAGC |
AS-NR-002-1-122 |
| 5016A | GGGGGTATAGCTCAG | CysGCA | AS-NR-002-1-123 |
| 5019A | GGTAGCGTGGCCGAGC | LeuTAG | AS-NR-002-1-124 |
| 5019B | GGTAGCGTGGCCGAGCGGTCTAAG | LeuTAG | AS-NR-002-1-125 |
| 5020/21A | GGTTCCATAGTGTA GGTTCCATGGTGTA |
ValTAC GlnCTG |
AS-NR-002-1-126 |
| 5020B | GGTTCCATAGTGTAGTGGTTATC | ValTAC | AS-NR-002-1-127 |
| 5021B | GGTTCCATGGTGTAATGG | GlnCTG | AS-NR-002-1-128 |
| 5022A | GTAGTCGTGGCCGA | SerTGA | AS-NR-002-1-129 |
| 5022B | GTAGTCGTGGCCGAGTGGTTAAGGC | SerTGA | AS-NR-002-1-130 |
| 5023B | GTCAGGATGGCCGAGCGGTCTAA | LeuCAG | AS-NR-002-1-131 |
| 5024A | GTTAAGATGGCAGA | LeuTAA | AS-NR-002-1-132 |
| 5026A | GTTTCCGTAGTGTAGTGG | ValCAC | AS-NR-002-1-133 |
| 5026/27B | GTTTCCGTAGTGTAGTGGTCATC GTTTCCGTAGTGTAGTGGTTATC |
ValAAC ValCAC ValAAC |
AS-NR-002-1-134 |
| 5028/29A | TCCCACATGGTCTAGCGG TCCCATATGGTCTAGCGG |
GluTTC GluTTC |
AS-NR-002-1-135 |
| 5028/29B | TCCCACATGGTCTAGCGGTTAGG TCCCATATGGTCTAGCGGTTAGG |
GluTTC GluTTC |
AS-NR-002-1-136 |
| 5032A | TCCTCGTTAGTATAGTGG | AspGTC | AS-NR-002-1-137 |
| 5032B | TCCTCGTTAGTATAGTGGTGAGT | AspGTC | AS-NR-002-1-138 |
| TRF21-26 | ACCAGAATGGCCGAGTGGTT | LeuTAA | AS-NR-002-1-139 |
| TRF63 | AGCAGAGTGGTGCAGTGG | MetCAT | AS-NR-002-1-140 |
| TRF23 | ACCCTGTGGTCTAGTGG | GluTTC | AS-NR-002-1-141 |
| TRF205 | CCCCTGGTGGTCTAGTGCTTAGGATTT | GluCTC | AS-NR-002-1-142 |
| TRF208 | CCCTGTGGTCTAGTGGTTAG | GluTTC | AS-NR-002-1-143 |
| TRF223 | CGCTCTTTGGTCTAGGGG | ProAGG | AS-NR-002-1-144 |
| TRF250 | CTCGTTAGTATAGTGGT | AspGTC | AS-NR-002-1-145 |
| TRF272/274 | GACCTCGTGGCGCAACGG GACCTCGTGGCGCAATGG |
TrpCCA TrpCCA |
AS-NR-002-1-146 |
| TRF273 | GACCTCGTGGCGCAACGGT | TrpCCA | AS-NR-002-1-147 |
| TRF293/294 | GCATGGGTGATTCAGTGGTAGAATTT GCATGGGTGGTTCAGTGGTAGAATTC |
GlyGCC GlyGCC |
AS-NR-002-1-148 |
| TRF305/306 307 |
GCCCCAGTGGCCTAATGGA GCCCCAGTGGCCTGATGGA GCCCCGGTGGCCTAATGGA |
ArgCCT ArgCCT ArgCCT |
AS-NR-002-1-149 |
| TRF308 | GCCCGACTACCTCAGTCGG | LysCTT | AS-NR-002-1-150 |
| TRF312 | GCCCGGATGATCCTCAGTGGT | SeCTCA | AS-NR-002-1-151 |
| TRF316 | GCCCTCTTAGCGCAGTGGG | MetCAT | AS-NR-002-1-152 |
| TRF318 | GCCGAAATAGCTCAGTTGG | PheGAA | AS-NR-002-1-153 |
| TRF320 | GCCGTGATCGTATAGTGGTTA | HisGTG | AS-NR-002-1-154 |
| TRF321 | GCCTCCTTAGCGCAG | MetCAT | AS-NR-002-1-155 |
| TRF322 | GCCTCGTTAGCGCAGTAGG | MetCAT | AS-NR-002-1-156 |
| TRF323/324 326 |
GCCTGGATAGCTCAGTCGG GCCTGGATAGCTCAGTTGG GCCTGGGTAGCTCAGTCGG |
LysTTT LysTTT LysTTT |
AS-NR-002-1-157 |
| TRF337-339 | GCGTTGGTGGTATAGTGGT | GlyTCC | AS-NR-002-1-158 |
| TRF347 | GCTTCTGTAGTGTAGTGG | ValCAC | AS-NR-002-1-159 |
| TRF351 | GGATTGGTGGTCCAGTGGTAGAATTC | ArgCCT | AS-NR-002-1-160 |
| TRF354 | GGCCCTATAGCTCAGGGG | ThrTGT | AS-NR-002-1-161 |
| TRF356/359 | GGCCGCGTGGCCTAATGGA GGCCGTGTGGCCTAATGGA |
ArgTCG ArgTCG |
AS-NR-002-1-162 |
| TRF368 | GGCTCCGTGGCGCAATGGA | ArgTCT | AS-NR-002-1-163 |
| TRF366 | GGCTCCATAGCTCAGTGGTTAGAGCA | ThrTGT | AS-NR-002-1-164 |
| TRF365 | GGCTCCATAGCTCAGGGGT | ThrTGT | AS-NR-002-1-165 |
| TRF373 | GGCTCTGTGGCGCAATGGA | ArgTCT | AS-NR-002-1-166 |
| TRF374 | GGCTCTGTGGCTTAGTTGGC | ThrCGT | AS-NR-002-1-167 |
| TRF375 | GGCTTCGTGGCTTAGCTGG | ThrAGT | AS-NR-002-1-168 |
| TRF393 | GGGGGCATAGCTCAGTGG | CysGCA | AS-NR-002-1-169 |
| TRF396 | GGGGGTATAGCTCAGCGGT | AlaAGC | AS-NR-002-1-170 |
| TRF417 | GGTAGCGTGGCCGAGTGGTCT | LeuTAG | AS-NR-002-1-171 |
| TRF457 | GTAGTCGTGGCCGAGTGG | SerAGA | AS-NR-002-1-172 |
| TRF460 | GTCACGGTGGCCGAGTGG | SerCGA | AS-NR-002-1-173 |
| TRF462 | GTCAGGATGGCCGAGCGGT | LeuCAG | AS-NR-002-1-174 |
| TRF463 | GTCAGGATGGCCGAGTGG | LeuCAA | AS-NR-002-1-175 |
| TRF466 468 469 471 472 473 |
GTCTCTGTGGCACAATCGGT GTCTCTGTGGCGCAATCGG GTCTCTGTGGCGCAATCGGT GTCTCTGTGGCGCCATCGGT GTCTCTGTGGCGTAGTCGGT GTCTCTGTGGTGCAATCGGT |
AsnGTT AsnGTT AsnGTT AsnGTT AsnGTT AsnGTT |
AS-NR-002-1-176 |
| TRF490 | GTTAAGATGGCAGAG | LeuTAA | AS-NR-002-1-177 |
| TRF492 | GTTAAGATGGCAGAGCCTGGT | LeuTAA | AS-NR-002-1-178 |
| TRF493 | GTTAAGATGGCATAGCCC | LeuTAA | AS-NR-002-1-179 |
| TRF511 | GTTTCCGTAGTGTAG | ValCAC | AS-NR-002-1-180 |
| TRF524 | TAGGATGTGGTGTGACAG | GlnTTG | AS-NR-002-1-181 |
| TRF533/534 | TCCCACATGGTCTAGCGGT TCCCATATGGTCTAGCGGT |
GluTTC GluTTC |
AS-NR-002-1-182 |
| TRF537 | TCCCCTTGGTCTAGTGGTTAGGATTC | GluCTC | AS-NR-002-1-183 |
| TRF546/547 | TCCTCGTTAGTATAGTGGT TCCTCATCAGTATAGTGGT |
AspGTC AspGTC |
AS-NR-002-1-184 |
| TRF550/551 | TCCTTGGTGGTCTAGTGGCTAG TCCTTGATGTCTAGTGGTTAG |
GluTTC GluCTC |
AS-NR-002-1-185 |
| House Keeping Gene | Catalog Number |
| RNU6-2 | AS-NR-002-1-186 |
| SNORD43 | AS-NR-002-1-187 |
| SNORD95 | AS-NR-002-1-188 |
| tRF ID in tRFdb | tRF Type | Catalog Number |
| 1003 | tRF-1 | AS-NR-002M-1-001 |
| 1006 | tRF-1 | AS-NR-002M-1-002 |
| 1008 | tRF-1 | AS-NR-002M-1-003 |
| 1009 | tRF-1 | AS-NR-002M-1-004 |
| 1010 | tRF-1 | AS-NR-002M-1-005 |
| 1015 | tRF-1 | AS-NR-002M-1-006 |
| 1016 | tRF-1 | AS-NR-002M-1-007 |
| 1019 | tRF-1 | AS-NR-002M-1-008 |
| 1020 | tRF-1 | AS-NR-002M-1-009 |
| 1026 | tRF-1 | AS-NR-002M-1-010 |
| 1035 | tRF-1 | AS-NR-002M-1-011 |
| 3001b | tRF-3 | AS-NR-002M-1-012 |
| 3002a/3035a | tRF-3 | AS-NR-002M-1-013 |
| 3003a | tRF-3 | AS-NR-002M-1-014 |
| 3004b | tRF-3 | AS-NR-002M-1-015 |
| 3005a | tRF-3 | AS-NR-002M-1-016 |
| 3006a | tRF-3 | AS-NR-002M-1-017 |
| 3009a | tRF-3 | AS-NR-002M-1-018 |
| 3009b | tRF-3 | AS-NR-002M-1-019 |
| 3010a | tRF-3 | AS-NR-002M-1-020 |
| 3010b | tRF-3 | AS-NR-002M-1-021 |
| 3011a | tRF-3 | AS-NR-002M-1-022 |
| 3011b | tRF-3 | AS-NR-002M-1-023 |
| 3012b | tRF-3 | AS-NR-002M-1-024 |
| 3017a | tRF-3 | AS-NR-002M-1-025 |
| 3017b | tRF-3 | AS-NR-002M-1-026 |
| 3019a | tRF-3 | AS-NR-002M-1-027 |
| 3019b/3020b | tRF-3 | AS-NR-002M-1-028 |
| 3021a | tRF-3 | AS-NR-002M-1-029 |
| 3023b | tRF-3 | AS-NR-002M-1-030 |
| 3024b/3046b | tRF-3 | AS-NR-002M-1-031 |
| 3025a | tRF-3 | AS-NR-002M-1-032 |
| 3025b | tRF-3 | AS-NR-002M-1-033 |
| 3026a | tRF-3 | AS-NR-002M-1-034 |
| 3027a | tRF-3 | AS-NR-002M-1-035 |
| 3028b/3029b | tRF-3 | AS-NR-002M-1-036 |
| 3028b/3029b/3040b/3045b | tRF-3 | AS-NR-002M-1-037 |
| 3029a | tRF-3 | AS-NR-002M-1-038 |
| 3030a | tRF-3 | AS-NR-002M-1-039 |
| 3031a | tRF-3 | AS-NR-002M-1-040 |
| 3031b | tRF-3 | AS-NR-002M-1-041 |
| 3032a | tRF-3 | AS-NR-002M-1-042 |
| 3032b | tRF-3 | AS-NR-002M-1-043 |
| 3033a | tRF-3 | AS-NR-002M-1-044 |
| 3033b | tRF-3 | AS-NR-002M-1-045 |
| 3034a | tRF-3 | AS-NR-002M-1-046 |
| 3036a | tRF-3 | AS-NR-002M-1-047 |
| 3036b/3037b | tRF-3 | AS-NR-002M-1-048 |
| 3038a | tRF-3 | AS-NR-002M-1-049 |
| 3038b | tRF-3 | AS-NR-002M-1-050 |
| 3039a | tRF-3 | AS-NR-002M-1-051 |
| 3041b/3042b | tRF-3 | AS-NR-002M-1-052 |
| 3043a | tRF-3 | AS-NR-002M-1-053 |
| 3043b | tRF-3 | AS-NR-002M-1-054 |
| 3044a | tRF-3 | AS-NR-002M-1-055 |
| 3044b | tRF-3 | AS-NR-002M-1-056 |
| 3045a | tRF-3 | AS-NR-002M-1-057 |
| 3046a | tRF-3 | AS-NR-002M-1-058 |
| 3047b | tRF-3 | AS-NR-002M-1-059 |
| 3048a | tRF-3 | AS-NR-002M-1-060 |
| 3050a | tRF-3 | AS-NR-002M-1-061 |
| 3051a | tRF-3 | AS-NR-002M-1-062 |
| 3052a | tRF-3 | AS-NR-002M-1-063 |
| 5001a/5001b/5010a | tRF-5 | AS-NR-002M-1-064 |
| 5002b/5004b | tRF-5 | AS-NR-002M-1-065 |
| 5005a/5006b | tRF-5 | AS-NR-002M-1-066 |
| 5005b | tRF-5 | AS-NR-002M-1-067 |
| 5006a | tRF-5 | AS-NR-002M-1-068 |
| 5006c | tRF-5 | AS-NR-002M-1-069 |
| 5007a | tRF-5 | AS-NR-002M-1-070 |
| 5009a | tRF-5 | AS-NR-002M-1-071 |
| 5009b | tRF-5 | AS-NR-002M-1-072 |
| 5011a | tRF-5 | AS-NR-002M-1-073 |
| 5011b/5012b | tRF-5 | AS-NR-002M-1-074 |
| 5012b | tRF-5 | AS-NR-002M-1-075 |
| 5013a | tRF-5 | AS-NR-002M-1-076 |
| 5013b | tRF-5 | AS-NR-002M-1-077 |
| 5013b/5017b/5018b | tRF-5 | AS-NR-002M-1-078 |
| 5014a | tRF-5 | AS-NR-002M-1-079 |
| 5014a/5015a | tRF-5 | AS-NR-002M-1-080 |
| 5014b | tRF-5 | AS-NR-002M-1-081 |
| 5016a | tRF-5 | AS-NR-002M-1-082 |
| 5019a | tRF-5 | AS-NR-002M-1-083 |
| 5019b | tRF-5 | AS-NR-002M-1-084 |
| 5020b/5021a | tRF-5 | AS-NR-002M-1-085 |
| 5022a | tRF-5 | AS-NR-002M-1-086 |
| 5022b | tRF-5 | AS-NR-002M-1-087 |
| 5023b/5024b | tRF-5 | AS-NR-002M-1-088 |
| U6 | Reference gene | AS-NR-002M-1-089 |
| miR-93-5p | Reference gene | AS-NR-002M-1-090 |
| 5S rRNA | Reference gene | AS-NR-002M-1-091 |

