Order Guide
Related Products
Related Services
Related Reviews
nrStar™ Functional LncRNA PCR Array (H/M)
• Well-known LncRNAs characterized for biological functions or disease association are comprehensively collected.
• Functional and disease LncRNAs are carefully categorized, annotated and cross-referenced.
• All the primer sets are fully validated in numerous sample types to meet the needs of biomarker discovery.
• Transcript-specific PCR primers enable unambiguous and accurate detection of each individual LncRNA isoforms.
We recommend using our rtStar™ First-Strand cDNA Synthesis Kit (AS-FS-001) along with Arraystar SYBR® Green Real-time qPCR Master Mix when running your nrStar™ Functional LncRNA PCR Arrays for optimal performance.
| Product Name | Catalog No. | Size | Price |
|---|---|---|---|
| nrStar™ Human Functional LncRNA PCR Array | AS-NR-004-1 | 384-well/ plate | $368.00 |
| nrStar™ Human Functional LncRNA PCR Array (Roche Light Cycler 480) | AS-NR-004-1-R | 384-well/ plate | $368.00 |
| nrStar™ Mouse Functional LncRNA PCR Array | AS-NR-004M-1 | 384-well (2*192) / plate | $443.00 |
| nrStar™ Mouse Functional LncRNA PCR Array (Roche Light Cycler 480) | AS-NR-004M-1-R | 384-well (2*192) / plate | $443.00 |
• Best coverage for known functional lncRNAs: Well-known LncRNAs characterized for biological functions or human disease association are comprehensively collected from the most updated public databases and landmark publications.
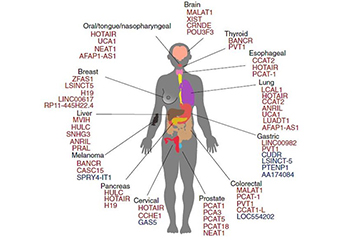
Figure 1. Some lncRNAs associated with cancers were collected from Bartonicek et al. 2016.
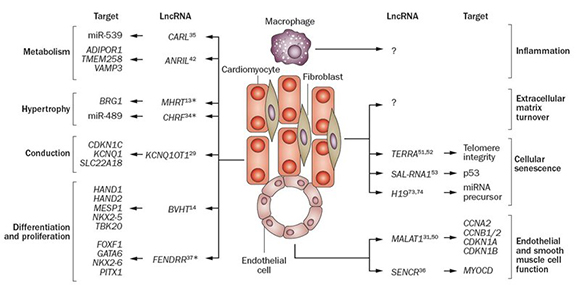
Figure 2. Some LncRNAs involved in cardiac pathways were collected from Devaux et al. 2015
• Comprehensive annotations: The LncRNAs that have been associated with diseases including cancers, cardiovascular diseases and neurodegenerative diseases are carefully categorized, annotated and cross-referenced.
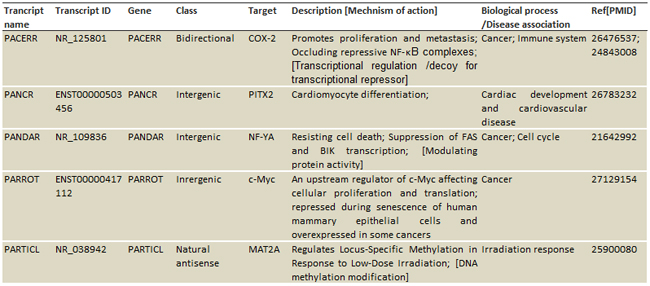
Table1. The annotated information includes:
Class based on genomic location: [enhancer, antisense, intergenic, intronic, bidirectional…]
Target mRNA genes
Mechanisms: [signal, decoy, scaffold, guide, ceRNA, miRNA sponge, splicing …]
Biological functions: [cell cycle, differentiation, methylation, immune, metabolism…]
Diseases: [cancers, cardiovascular, neurodegenerative, kidney, diabetes, syndromes…]
Annotation Example 1 – Cancer-associated lncRNA NBAT-1
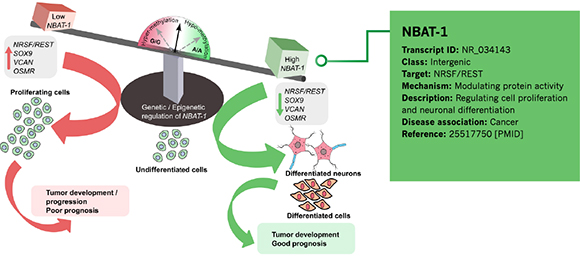
Figure 3. Information of NBAT-1 annotated by Arraystar is shown in the green rectangular area.
Annotation Example 2 – LncRNA Mhrt in cardiovascular diseases
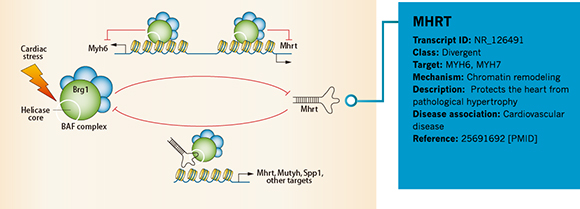
Figure 4. Information of MHRT annotated by Arraystar is shown in the blue rectangular area.
Annotation Example 3 – LncRNA BACE1-AS in neurodegenerative diseases
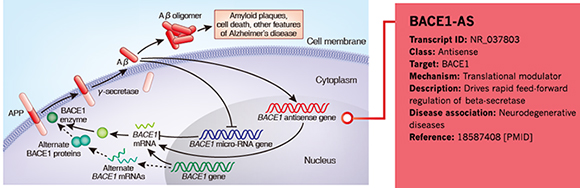
Figure 5. Information of BACE1-AS annotated by Arraystar is shown in the red rectangular area.
• A powerful tool for biomarker screening and validation: All the primer sets are optimized and fully validated experimentally using numerous sample types, to meet the needs of biomarker discovery.
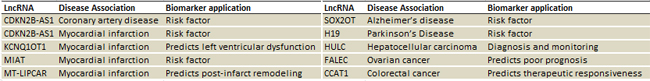
Figure 6. A snippet of the lncRNAs on the PCR array with published biomarker potentials and utilities.
• Transcript-specific detection: Transcript-specific PCR primers are designed to target the specific exon or splice junction sites, for unambiguous and accurate detection of each individual LncRNA isoforms.
• Most reliable and accurate quantification: Each lncRNA is quantified at high accuracy and data quality, by using external controls (RNA Spike-In, and positive PCR control), Genomic DNA Control and six internal housekeeping genes. Normalization is performed by geometric means of multiple reference genes selected by experimentally validated algorithm, instead of a single reference gene.
• Fast and easy: Ready-to-use PCR array in a 384-well plate format. The entire process can be completed in less than 4 hours.
Human Functional LncRNA List
| Cancer: 7SK, AA174084, AB073614, ABHD11-AS1, ACTA2-AS1, ADAMTS9-AS2, AF339813, AFAP1-AS1, AIRN-1, AIRN-2, AK095147, AP5M1, APOC1P1, ARA-1, ARA-2, ATG9B-1, ATG9B-2, BANCR, BCAR4-1, BCAR4-2, BGLT3, BLACAT1, BOK-AS1, C5orf66-AS1, CASC11-1, CASC11-2, CASC2-1, CASC2-2, CASC9, CBR3-AS1-1, CBR3-AS1-2, CCAT1, CCAT2, CCDC26, CCEPR, CRNDE, CTB-89H12.4, CTBP1-AS, DLEU1-1, DLEU1-2, DNM3OS-1, DNM3OS-2, DRAIC (LOC145837), DSCAM-AS1-1, DSCAM-AS1-2, EGFR-AS1, EGOT, ENST00000434223, ENST00000456816, ENST00000480739, EPB41L4A-AS1, EPB41L4A-AS2, EWSAT1, FALEC, FAS-AS1 (SAF), FER1L4, FEZF1-AS1, FGF14-AS2, FLG-AS1-1, FLG-AS1-2, FOXCUT, FTX, GACAT1-1, GACAT1-2, GACAT2, GACAT3, GAPLINC-1, GAPLINC-2, GAS5, GAS8-AS1, GATA6-AS1, GHET1, H19-1, H19-2, HAGLR, HAND2-AS1, HCP5, HIF1A-AS1, HIF1A-AS2, HNF1A-AS1, HOTAIR-1, HOTAIR-2, HOTAIR-3, HOTAIRM1-2, HOTTIP, HOXA-AS2 , HULC, KCNQ1OT1, KRASP1, KRT18P55, KRT7-AS, L1PA16, LINC00032, LINC00152-1, LINC00152-2, LINC00152-3, LINC00312, LINC00467, LINC00473, LINC00668, LINC00673, LINC00857, LINC00901 , LINC00951, Linc00963, LINC00970, LINC01133, LINC01315-1, LINC01315-2, LINC01630-1, LINC01630-2, LINC-PINT, Linc-POU3F3-1, Linc-POU3F3-2, lincRNA-p21, lincRNA-RoR, lncRNA-CTD903, lncRNA-HEIH, LOC100130476, LOC100507661 , LOC101054525 , LOC389332, LSINCT5, LUCAT1, LUNAR1, LUST, MACROD2-AS1-1, MACROD2-AS1-2, MEG3-1, MEG3-2, MER11C, MIAT, MINA, MIR100HG, MIR155HG, MIR17HG, MIR31HG, MT1JP, MYCNOS-1, MYCNOS-2, MYCNUT, NAMA-1, NAMA-2, NBAT-1, NDM29, NEAT1-1, NEAT1-2, NKILA, NPTN-IT1, OR3A4P, ORAOV1, PACERR, PANDAR, PARROT, PAX8-AS1-1, PAX8-AS1-2, PCA3-1, PCA3-2, PCAT-1, PCAT18, PCAT29, PCAT6-1, PCAT6-2, PCBP2-OT1, PCGEM1, PGM5-AS1-1, PGM5-AS1-2, PICSAR, PINC, POU6F2-AS2, PRNCR1, PSF inhibiting RNA, PTCSC1, PTCSC3, PTENP1, RGMB-AS1, RMEL3, RUNX1-IT1, SAMMSON , SBF2-AS1, SCHLAP1, SNHG1, SNHG15, SNHG16, SNHG20, SNHG5, SOX2OT, ST7-AS1, ST7-AS2-1, ST7-AS2-2, ST7-OT3, ST7-OT4, SUMO1P3, TARID, TCL6, TDRG1, TERC, TIE1-AS, TRERNA1, TRIM52-AS1-1, TRIM52-AS1-2, TUG1-1, TUG1-2, TUSC7 , TUSC8, U79277, uc.338, uc.73A(P), UCA1, UFC1 lincRNA , VLDLR-AS1, WSPAR, WT1-AS-1, WT1-AS-2, WT1-AS-3, X91348, Yiya, ZEB2-AS1, ZFAS1-1, ZNF582-AS1-1, ZNF582-AS1-2, ZNF582-AS1-3 |
| Neurodegenerative diseases: 17A, 51A, anti-NOS2A, ATXN8OS , BACE1-AS, BDNF-AS-1, BDNF-AS-2, CDKN2B-AS1, DGCR5, DLX6-AS1, DPY19L2P2-1, DPY19L2P2-2, GDNF-AS1 -1, H19-1, H19-2, HAR1A, HAR1B (HAR1R), HTT-AS, LINC00299, LINC00599, LINC01262, lincRNA-p21, MALAT1, MEG3-1, MEG3-2, MIAT, NAT-RAD18, NEAT1-1, NEAT1-2, PINK1-AS, PNKY, RMST, SCAANT1, SNHG1, SNHG3, SOX2OT, TUG1-1, TUG1-2, TUNA-1, TUNA-2, U1 spliceosomal lncRNA, UCH1LAS |
| Cardiovascular diseases: 7SK, ALIEN, ATG9B-1, ATG9B-2, CARMEN-1, CDKN2B-AS1, EMX2OS, FENDRR, FGF10-AS1, GAS5, H19-1, H19-2, HAS2-AS1, HIF1A-AS1, HIF1A-AS2, HOTAIR-1, HOTAIR-2, HOTAIR-3, KCNQ1OT1, LINC00323, lincRNA-p21, LOC100129973, LOC100507537, MHRT, MIAT, MIR222HG, NONHSAT073641, NONHSAT112178, Novlnc35, Novlnc44, Novlnc76, NPPA-AS1, NRON, PANCR, PUNISHER, PVT1, SALRNA1, SENCR, SMILR, TERMINATOR, TIE1-AS, TUG1-1, TUG1-2, UCA1 |
| Kidney diseases: ENST00000456816, GAS5, H19-1, H19-2, HIF1A-AS1, HIF1A-AS2, HOTAIR-1, HOTAIR-2, HOTAIR-3, KCNQ1OT1, LOC389332, MEG3-1, MEG3-2, PVT1, RP11-354P17.15-001, TapSAKI, X91348 |
| Diabetes: CDKN2B-AS1, HI-LNC25-1, HYMAI, IGF2-AS-1, LINC00271, MEG3-1, MEG3-2, NONRATT021972, PDZRN3-AS1, PVT1 |
| Immune system: GATA3-AS1-1, GATA3-AS1-2, GATA3-AS1-3, IFNG-AS1-1, IFNG-AS1-2, Linc-DC , lincRNA-EPS, M18204, MAFTRR, NRAV, NRIR, NTT, PACERR, RHOXF1P1, THRIL |
| Cell Cycle: CCND1 associated ncRNAs, CDKN2B-AS1, H19-1, H19-2, HULC, KCNQ1OT1, lincRNA-p21, lincRNA-RoR, lncRNA-HEIH, PANDAR, TUSC7 |
| Lipid metabolism and adipogenesis: APOA1-AS, HOTAIR-1, HOTAIR-2, HOTAIR-3, HULC, NEAT1-1, NEAT1-2, RP1-13D10.2 |
Mouse Functional LncRNA List
| Cancer: 9530018H14Rik; Airn-1; Airn-2; Airn-3; C130071C03Rik-1; C130071C03Rik-2; Dreh; Esrp2-as-1; Esrp2-as-2; Esrp2-as-3; H19; Hottip-1; Hottip-2; Lncpint-1; Lncpint-2; Lncpint-3; lncRNA-Smad7-1; lncRNA-Smad7-2; lncRNA-Smad7-3; Malat1; Meg3-1; Meg3-2; Mira; Pinc; Trp53cor1 |
| Cardiac development & cardiovascular diseases: 4930503E24Rik; AK028007; AK038798; AK044955; AK139328; AK143294; AK143693; AK153778; Atp2a2; Bvht; C2dat1; CAIF; Chaer; Cyp4b1-ps2; Chast; Dworf; Fendrr-1; Fendrr-2; Gm12919; Gm13865; Gm14155; Gm15054; Gm15834; Gm32592; Gm6644; Gm6768; LeXis-1; LeXis-2; Meg3-1; Meg3-2; Mhrt; Mirt1; Sghrt-1; Sghrt-2; Srsf9; Tdpx-ps1; TK99129; Uph; Wt1os |
| Diabete: LincRNA-Gm4419-1; LincRNA-Gm4419-2; Meg3-1; Meg3-2; Pluto; Rian |
| Embryonic Stem Cell Pluripotency & Differentiation: 1700007L15Rik; Digit; Evx1as; G730013B05Rik-1; G730013B05Rik-2; Gm16845-1; Gm16845-2; Gm2694-1; Gm2694-2; Halr1; linc1242; linc1257-1; linc1257-2; linc1368; Panct1; Snhg3 |
| Genomic imprinting: Kcnq1ot1; Nctc1; Nespas |
| Immune system development & immune response & immune disease: Gas5; Lnc13; lncKdm2b; lncRNA-ACOD1; lncRNA-Nfkb2-1; lncRNA-Nfkb2-2; lnc-Smad3; Malat1; Mir142hg; Mirt2; Morrbid-1; Morrbid-2; Morrbid-3; Ptgs2os2; Rmrp; Rroid |
| Kidney diseases: np_17856, np_5318, Xist-1; Xist-2 |
| Liver diseases: Airn-1; Airn-2; Airn-3; APOA4-AS; lnc-LFAR1 |
| Lipid metabolism & adipogenesis & adipocyte differentiation: C730029A08Rik; C730036E19Rik; Gm15050; Gm16551 |
| Neuronal development & Differentiation & Diseases: 4930570G19Rik-1; 4930570G19Rik-2; Abhd11os; AK133808; Dalir; Dleu2; Dlx1as; Egr2-AS; Gm30731; lncOL1; lncOL1; Miat-1; Miat-2; Nkx2-2os; Paupar; Rmst; Six3os1-1; Six3os1-2; Six3os1-3; Six3os1-4; Six3os1-5; Six3os1-6; Sox2ot; Uchl1os |
| rRNA metabolism: 5S-OT, PAPAS |
| Spermatogonial stem cells survival & spermatogenesis & infertility: Gfra1; Hm629797; lncRNA033862; Neat1-1; Neat1-2 |
| Skeletal & muscle development: LOC105246506; lnc-31; Msx1os; Munc; Myhas; Yam-1 |
| X inactivation: Firre; Ftx-1; Ftx-2; Jpx; Rr18; Tsix; Xist-1; Xist-2 |
| Other biological functions & diseases: 6430411K18Rik; AK081227; alncRNA-EC7; CAIF; Egr2-AS; Hotair; Hoxaas3; lncLGR; Mdrl; Mir124a-1hg; Neat1-1; Neat1-2; Rubie; Smn-AS1; Snhg5; Tog; Ttc39aos1; Tug1-1; Tug1-2; Tug1-3; Vax2os-1; Vax2os-2 |
Suffix (-1, -2, -3 …) represents different transcript isoforms of the lncRNA genes.

Figure 1. Workflow of nrStar™ Functional LncRNA PCR Array
Compatible qPCR Instruments Equipped with a 384-well Format Block:
ABI ViiA™ 7
ABI 7500 & ABI 7500 FAST
ABI 7900HT
ABI QuantStudio™ 5 Real-Time PCR system
ABI QuantStudio™ 6 Flex Real-Time PCR system
ABI QuantStudio™ 7 Flex Real-Time PCR system
ABI QuantStudio™ 12K Flex Real-Time PCR System
Bio-Rad CFX384
Bio-Rad iCycler & iQ Real-Time PCR Systems
Eppendorf Realplex
Stratagene Mx3000
Roche LightCycler 480
The role of mechanical strain-induced HOTAIR lncRNA downregulation in the pathophysiology of rheumatoid arthritis. Meier F, et al. Annals of the Rheumatic Diseases, 2024
Exosomal long non-coding RNA AU020206 alleviates macrophage pyroptosis in atherosclerosis by suppressing CEBPB-mediated NLRP3 transcription. Zhang L, et al. Experimental Cell Research, 2024
Long non-coding RNA small nucleolar RNA host gene 1 alleviates the progression of recurrent spontaneous abortion via the microRNA-183-5p/ZEB2 axis. Mo Y, et al. Reproductive Biology, 2022
CAD increases the long noncoding RNA PUNISHER in small extracellular vesicles and regulates endothelial cell function via vesicular shuttling. Hosen M R, et al. Molecular Therapy. Nucleic Acids, 2021
Tumor-associated mesenchymal stem cells promote hepatocellular carcinoma metastasis via a DNM3OS/KDM6B/TIAM1 axis. Wang W, et al. Cancer Letters, 2021
Mesenchymal stem cell-derived exosomes block malignant behaviors of hepatocellular carcinoma stem cells through a lncRNA C5orf66-AS1/microRNA-127-3p/DUSP1/ERK axis. Gu H, et al. Human Cell, 2021
Involvement of the lncRNA AFAP1-AS1/microRNA-195/E2F3 axis in proliferation and migration of enteric neural crest stem cells of Hirschsprung’s disease. Pan W, et al. Experimental Physiology, 2020
LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Gao H, et al. International journal of biological macromolecules, 2018
Saturated fatty acid alters embryonic cortical neurogenesis through modulation of gene expression in neural stem cells. Ardah M T, et al. Journal of Nutritional Biochemistry, 2018


